
CongBao Kang
Agency for Science, Technology and Research (A*STAR), Singapore
Title: Structural and dynamic studies of DENV and ZIKV proteases and its insight into inhibitor design
Biography
Biography: CongBao Kang
Abstract
Dengue virus (DENV and Zika virus (ZIKV) belong to Flaviviridae genus which contains important human pathogens. DENV affects people living in tropical and subtropical regions. DENV infection can cause serious diseases such as dengue fever. ZIKV has drawn worldwide attention because of the outbreak in 2015. Viral genome of a flavivirus encodes a polyprotein that can be processed into structural and non-structural (NS) proteins by both host and viral proteases. Viral protease is a two-component serine protease formed by a cofactor region (~40 aa) from NS2B and a protease region (~170 aa) from NS3. The NS2B-NS3 protease of DENV or ZIKV is a validated target because of their function in maturation of viral proteins. Structural studies have been conducted for both DENV and ZIKV proteases. For DENV, previous studies have demonstrated that the free protease adopts an open conformation in which the C-terminal part of the NS2B cofactor region stays away from the active site. In the presence of an inhibitor, DENV protease forms a closed conformation in which the C-terminal region of NS2B forms part of the active site and interacts with the inhibitor. Our NMR study reveals that an unlinked DENV protease adopts the closed conformation in solution. Based on the knowledge on DENV protease, several constructs were made for ZIKV protease. Structural studies demonstrated that ZIKV protease adopts the closed conformation in the absence and presence of an inhibitor or substrate. The linker or enzymatic cleavage site present between NS2B and NS3 may affect inhibitor to interact with the active site. Our accumulated studies have shown that the unlinked protease construct can be used for studying protease-inhibitor interactions. We have demonstrated that the unlinked ZIKV protease interacts with different types of inhibitors. Our studies will be helpful for structure-based inhibitor design against both ZIKV and DENV proteases.
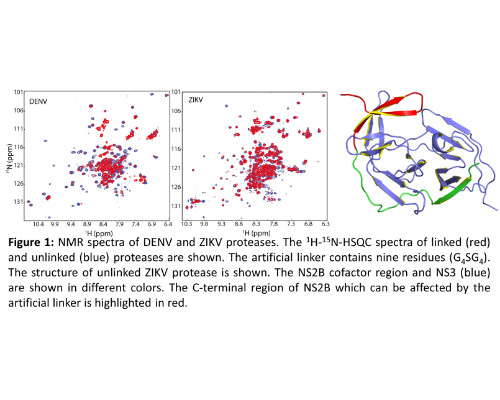
References:
- Zhang Z, Li Y, Loh YR, Phoo WW, Hung AW, Kang C*, Luo D* (2016) Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science, 354(6319):1597-1600.
- W. W. Phoo, Y. Li, Z. Z. Zhang, M. Y. Lee, Y. Loh, Y. B. Tan, E. Y. Ng, J. Lescar, C. Kang*, D. Luo* (2016) Structure of NS2B-NS3 Protease from Zika Virus after Self-cleavage. Nature communications, 7:13410.
- Y. Li, YL Wong, M.Y. Lee, Q. Li, J. Lescar, P.Y. Shi, C. Kang*, (2016) Secondary structure and membrane topology of the full length Dengue NS4B in micelles. Angewandte Chemie Int Ed Engl, 55(39):12068-72
- Y. Li, Q. Li, Y.L. Wong, L. Liew, and C.B. Kang,*(2015) Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. BBA-Biomembranes. 1848: 2244-2252.
- Zou J, Xie X., Lee LT., Chandrasekaran R., Reynaud A., Yap L., Wang Q., Dong H., Kang C.B., Yuan Z., Lescar J., and Shi P. (2014) Dimerization of Flavivirus NS4B Protein. Journal of Virology. 88(6): 3379-3391.
