
Fabio C. L. Almeida
Federal University of Rio de Janeiro, Brazil
Title: Role of conformational equilibrium in molecular recognition and capsid assembly: the case of flavivirus capsid proteins
Biography
Biography: Fabio C. L. Almeida
Abstract
Proteins are dynamic entities able to move in a wide range of timescales that goes from picoseconds to seconds. Motions that occur in microseconds to seconds define biologically relevant events that are frequently involved in binding, allostery and catalysis1,2. In our laboratory, we used relaxation parameter, relaxation dispersion experiments and molecular dynamic simulation to correlate conformational equilibrium with molecular recognition and catalysis.
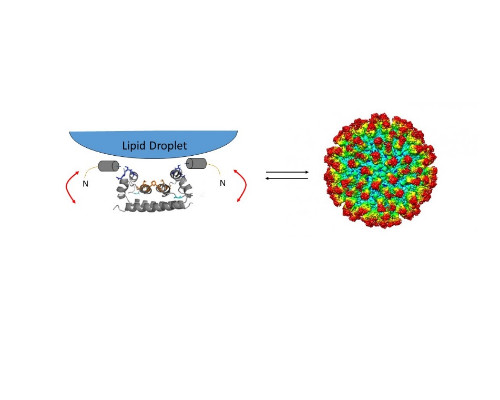
Dengue and Zika are major arthropod-borne human viral disease, for which no specific treatment is available. The flavivirus capsid protein is the trigger of virus assembly. Capsid proteins are located at the cytoplasm bound to lipid droplets (LD). Binding to LDs are essential for virus assembly3,4. We previously showed that the positively charged N-terminal region of Dengue virus capsid protein prompts the interaction with negatively charged LDs, after which a conformational rearrangement enables the access of the central hydrophobic patch to the LD surface5. We also showed the participation of the intrinsically disordered region in binding and possible regulation of capsid assembly6.
We probed the structure and dynamics of Dengue virus and Zika virus capsid proteins (DENVC and ZkC) by nuclear magnetic resonance. They bind lipid droplets (LD) in the cytoplasm, which mediates virus assembly in an unknown way. We showed that the dynamics of the capsid protein is intrinsically involved in the mechanism of LD and RNA binding and virus assembly. We also measured binding to nucleic acids and probed the assembly using small angle x-ray scattering and negative staining electron microscopy. The understanding of the participation of the intrinsically disordered N-terminal region and its dynamics helped us propose a mechanism for Dengue and Zika virus assembly and to develop a peptide with the potential to block virus assembly.
ACKNOLEDGEMENTS: FAPERJ, CAPES, CNPq, INBEB-CNPq.
References:
- Iqbal, A., Moraes, A. H., Valente, A. P. & Almeida, F. C. L. Structures of the reduced and oxidized state of the mutant D24A of yeast thioredoxin 1: insights into the mechanism for the closing of the water cavity. J. Biomol. NMR 63, 417–423 (2015).
- de Paula, V. S., Razzera, G., Barreto-Bergter, E., Almeida, F. C. L. & Valente, A. P. Portrayal of complex dynamic properties of sugarcane defensin 5 by NMR: multiple motions associated with membrane interaction. Structure 19, 26–36 (2011).
- Samsa, M. M. et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5, e1000632 (2009).
- Faustino, A. F. et al. Dengue virus capsid protein interacts specifically with very low-density lipoproteins. Nanomedicine 10, 247–55 (2014).
- Martins, I. C. et al. The disordered N-terminal region of dengue virus capsid protein contains a lipid droplet-binding motif. Biochem. J. 444, 405–415 (2012).
