
Beat Vögeli
University of Colorado at Denver, USA
Title: Functional protein conformation networks probed by NMR nanorulers
Biography
Biography: Beat Vögeli
Abstract
The function of a protein is tightly connected to its conformational network. Often, subtle differences distinguish interchanging states with distinct properties. One major challenge in structural biology is a sufficiently complete description of the structural landscape and the exchange dynamics between structural states at atomic resolution. We have replaced the standard NMR structure determination by an approach that generates multi-state ensembles from a dense network of tight averaged distance restraints derived from exact measurements of nuclear Overhauser enhancements (eNOEs) [Vögeli 2014, Vögeli et al. 2016].
Here, we present the identification of conformational networks harbored by the two human cis/trans isomerases cyclophilin A and Pin1 using the ‘nanorulers’ provided by eNOEs.
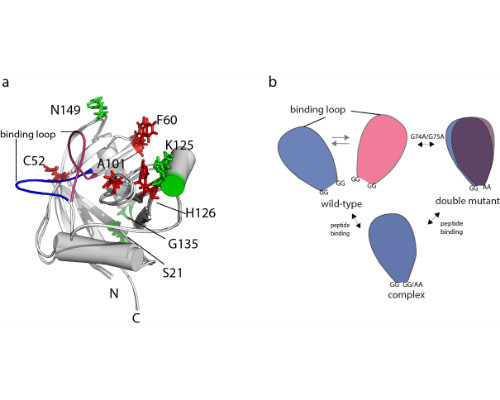
We have previously presented an eNOE-based ensemble description of cyclophilin that reveals the presence of a closed and an open state, the latter of which preorganizes the catalytic site for catalysis [Chi et al. 2015]. Based on this finding, we demonstrate here a ligand-selective change of the binding affinity to the active site by tuning of the dynamics of a highly flexible loop [Vögeli et al. 2016]. We show that the binding affinity is increased upon substitution of double glycines to alanines at either of the hinge regions of a loop. The equilibrium distribution is shifted towards more binding-competent conformations.
Comparison of the eNOE-based ensembles of the free and ligand-bound WW domain of Pin1 reveals a conformational network that extends into the interface formed with the enzymatically active PPIase domain. This finding may offer an atomic-picture explanation for the previously discovered communication between the two domains [Peng 2015].
References:
- Vögeli B, Kazemi S, Güntert P, Riek R (2012) Spatial elucidation of motion in proteins by ensemble-based structure calculation using exact NOEs. Nat Struct Mol Biol 19:1053-1057.
- Vögeli B, Olsson S, Güntert P, Riek R (2016) The exact NOE as an alternative in ensemble structure determination. Biophys J 110:113-126.
- Chi C, Vögeli B, Bibow S, Güntert P, Riek R (2015) A structural ensemble for the enzyme cyclophilin reveals an orchestrated mode of action at atomic resolution. Angew Chem Int Ed Engl 54:11657-11661.
- Vögeli B, Bibow S, Chi C (2016) Enzyme selectivity fine-tuned through dynamic control of a loop. Angew Chem Int Ed Engl 55:3096-30100.
- Peng J (2015) Investigating dynamic interdomain allostery in Pin1. Biophys Rev 7:239-249
