
Matthias Buck
Case Western Reserve University, USA
Title: Solution NMR relaxation and µs Molecular Dynamics Simulations of Dynamic Protein-Protein and Protein-Membrane Complexes
Biography
Biography: Matthias Buck
Abstract
It is now recognized that protein-protein interactions in solution are often dynamic, especially if the binding affinities are only moderately strong. Dynamics originate in part from the interconversion between structures of the protein complex, e.g. one bound state that is in equilibrium with one or several alternate configurations. We determined the structure of such a complex using NMR restraints [1] and saw the transitions between different configurations in microsecond length all-atom molecular dynamics simulations [2]. Recently, we also studied the dissociation process of mutant complexes which had a weakened primary interaction interface. Those simulations suggested that there is no single dissociation pathway, but that the separation first involves transitions to binding interfaces with fewer/weaker contacts [3].
Comparison is made between experimental NMR relaxation measurements on the ps-ns as well as µs-ms timescale with the microsecond all atom simulations, also in the context of new simulations of the protein association process. The functional significance of the protein complex alternate states and their dynamics are discussed.
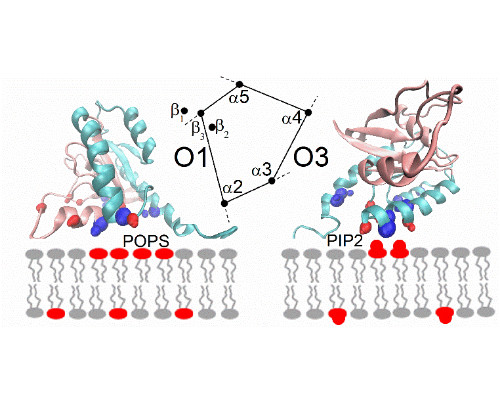
In a second part of the presentation, we consider a second system involving transient interactions; this time between K-Ras and the lipid bilayer of the plasma membrane [4]. Our recent simulations the full length GTPase at different membranes reveal the underlying rules of the interactions, emphasizing electrostatic contacts but also protein topology [5]. Again, simulations are compared with NMR experiments, carried out at model systems for the membrane.
References:
- Lee, HJ, Hota, P.K, Chugha, P., Miao, H., Zhang, L, Kim, SJ, Alviani, R.S, Stetzig, L., Wang, B. and Buck, M. (2012) “NMR structure of a hetero-dimeric SAM: SAM complex: Characterization and manipulation of EphA2 binding reveal new cellular functions of SHIP2”. Structure 20, 41-55.
- Zhang, L., and Buck, M. (2013) "Molecular Simulations of a Dynamic Protein Complex: Role of Salt-Bridges and Polar Interactions in Configurational Transitions" Biophys. J. 105, 2412-2417
- Zhang, L., Borthakur, S. and Buck, M. (2016) "Dissociation of a Dynamic Protein Complex studied by All Atom Molecular Simulation" Biophys. J., 110, 877-886.
- Li, Z., Cao, S., & Buck, M. (2016) “K-Ras at an Anionic Membrane: Orientation, orientation...orientation. Insights from recent simulation and experimental work”. New&Notable in Biophys. J. 110, 1033-1035.
- Li, Z. & Buck, M. (2017) “Computational Modelling Reveals Signaling Lipids Modulate the Orientation of K-Ras4A at the Membrane Reflecting Protein Topology.” Structure, revision submitted.
