
Bryan Kolaczkowski
University of Florida, USA
Title: An unbiased view of functional shifts and co-evolution across large protein families: Integrating computational and experimental approaches
Biography
Biography: Bryan Kolaczkowski
Abstract
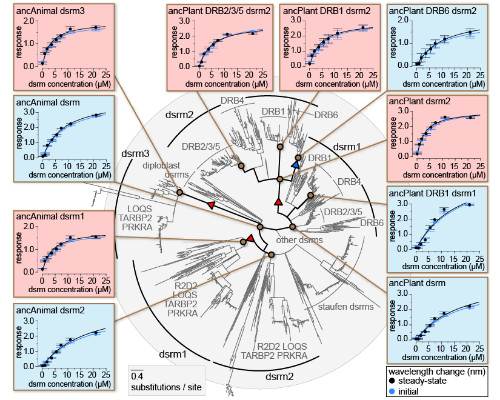
Determining the evolutionary forces driving transitions in protein function, the structural mechanisms through which functions change and the effects on cellular signaling is central to molecular evolution. These questions are commonly examined using computational and statistical approaches. More recently, ancestral sequence resurrection (ASR) has combined inference of ancient protein sequences with experimental functional characterization. However, the reliance of these studies on low-throughput experiments limits them to examining a relatively small number of hypotheses. Currently, we have little direct, unbiased information about how protein function may change across large phylogenies. Here we develop an approach integrating large-scale ancestral sequence reconstruction with structural modeling, molecular dynamics, function prediction and experimental validation to characterize molecular-functional shifts and their structural bases across protein families with 1000s of sequences. We apply this approach to study two families of RNA-binding proteins spanning animal and plant lineages. We identify discrete shifts in protein-ligand affinities and long-term changes in function occurring across multiple nodes. Our results suggest that changes in protein function may not always be associated with gene duplication or major speciation events. The identification of functional 'flip flopping' - repeated transitions among a small number of functional states through different structural mechanisms - also supports the view that protein function may be highly evolutionarily labile. We characterize co-evolution between RNA binding proteins and their signaling partners, suggesting that co-evolutionary processes may be common, even when large shifts in molecular function are rare. We suggest that an 'unbiased' view of protein functional evolution may reveal new information about how protein families evolve when we aren't looking.
References:
- Pugh C, Kolaczkowski O, Manny A, Korithoski B, Kolaczkowski B (2016) Resurrecting the ancestral structural dynamics of an antiviral immune receptor: Adaptive binding pocket reorganization repeatedly shifts RNA preference. BMC Evolutionary Biology 16(1):241.
- Dias R, Kolaczkowski B (2015) Different combinations of atomic interactions predict protein-small molecule and protein-DNA/RNA affinities with similar accuracy. Proteins 83(11):2100-2114.
- Korithoski B, Kolaczkowski O, Mukherjee K, Kola R, Earl C, Kolaczkowski B (2015) Evolution of a novel antiviral immune signaling interaction by partial-gene duplication. PLoS One 10(9):e0137276.
- Mukherjee K, Korithoski B, Kolaczkowski B (2014) Ancient origins of vertebrate-specific innate antiviral immunity. Molecular Biology and Evolution 31(1):140-153.
- Mukherjee K, Campos H, Kolaczkowski B (2013) Evolution of animal and plant Dicers: Early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Molecular Biology and Evolution 30(3):627-641.
